Up to exabytes of data are potentially available from public sources but the biomedical sciences remain largely siloed into well-demarcated strata. In fact, the hierarchical organization of biological complexity can be represented as a multi-layered chart, in which each layer represents a domain of knowledge.
Unfortunately, little if any integration is realized across disciplines either experimentally, analytically or conceptually. We argue that integration of information from seemingly disparate disciplines such as genetics, pharmacology, physiology, and cellular biology can facilitate the inference of logical, and biologically plausible novel relationships. Indeed, if enough information is connected via biologically relevant paths into a giant graph, new knowledge will be produced as an emergent property of such an ensemble.
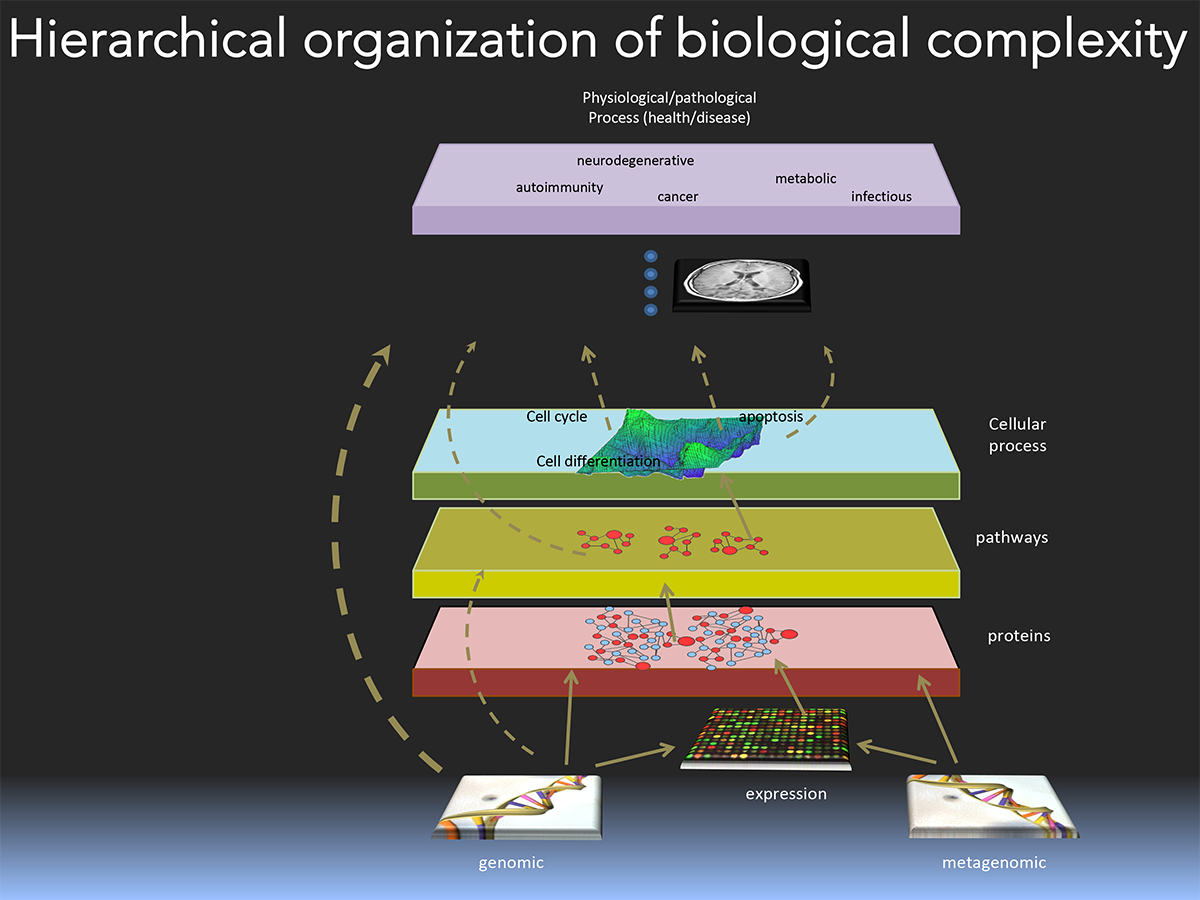
While integration and mining of large public datasets could prove a powerful approach to novel discoveries, these databases are often built using different architectures and standards, which hampers their straightforward integration
To tackle this problem, we created a scalable precision medicine open knowledge engine (SPOKE)—a giant heterogeneous graph currently boasting more than 3 million nodes and 5 million edges. SPOKE can be utilized to prioritize new uses for existing drugs (drug repurposing), to predict molecular targets of compounds and to create individualized patient profiles from electronic health records, among other applications. Current SPOKE projects at the Baranzini lab include:
Drug Repurposing for Progressive MS
Our lab is a partner in the BRAVEinMS consortium, a Progressive MS Alliance (PMSA)-sponsored collaboration with the goal of developing an effective therapeutic for progressive forms of MS using a drug repurposing approach.
Embedding of electronic health records (EHR) for precision medicine: We are developing approaches to integrate individual-specific medical information (from EHR) with population-specific knowledge (from SPOKE). The result of these approaches is a detailed “health barcode”, a map of the most biologically relevant variables within SPOKE for a particular patient, at a particular point in time. These barcodes can be utilized to group patients with similar characteristics, predict responders to therapeutic drugs, predict outcomes, understand mechanism of action of therapeutic drugs and more.
Scalable Precision Medicine Open Knowledge Engine
We are proud to commemerate nearly a decade of work on this project!
In 2015, we proposed a method to identify disease associated genes from edge prediction in hetergenous graphs that stored relational information between 18 different node types (including genes, diseases, tissues, pathophyiologices, etc.). Since that work, SPOKE has been dramatically expanded and utlized to study a wide variety of topics ranging from extraterrestrial diseases in spaceflown mice to early detection of Parkinson's disease.
We are excited to continue actively developing this project. Some of the many recent expansions of SPOKE include:
- We are actively increasing the number of databases that it relies on and hope to add many new node types ranging from Foods to Protein Structures
- We are extending the technical representations of graph
We continue towards our goal of developing a tool which can understand deep human biology

Try it yourself: SPOKE Explorer
The Scalable Precision-medicine Oriented Knowledge Engine (SPOKE) is a comprehensive biomedical knowledge graph connecting a wealth of information from basic molecular research, clinical insights, and many other databases. The SPOKE Neighborhood Explorer tool allows anyone to interact with the knowledge graph in a hypothesis-driven manner and browse connections between genes, drugs, diseases and more. Recently, the SPOKE team added Sars-CoV-2 data from the Krogan Lab's work examining the viral proteins. Pre-loaded queries in the Neighborhood Explorer let you explore the viral-human protein interactions and how they are connected to other data elements within SPOKE. Their team is using SPOKE to inform candidates for drug repurposing, and also in combination with EHRs to identify pre-existing conditions that can put people at higher risk of hospitalization
We are constantly adding more databases to SPOKE in order to enhance and its usefulness and broaden the scope of problems that could be tackled.
Additional SPOKE projects are underway at the Keiser (UCSF), Sui Huang (Institute for Systems Biology), Shivshankar Sundaram (LLNL) and Ramajrishnan Kannan (ORNL) labs.
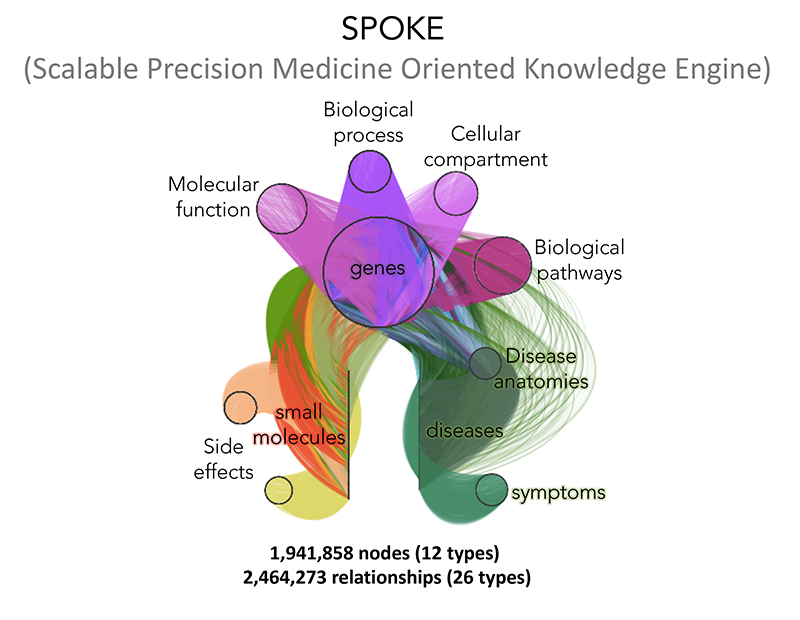
Read more by reviewing our recent publications on SPOKE below:
- The scalable precision medicine open knowledge engine (SPOKE): A massive knowledge graph of biomedical information
- Time-aware Embeddings of Clinical Data using a Knowledge Graph.
- A biomedical open knowledge network harnesses the power of AI to understand deep human biology.
- Embedding electronic health records onto a knowledge network recognizes prodromal features of multiple sclerosis and predicts diagnosis.